Paragraph 1: One thing I learned from this activity is that all the different types of genes are much more spread out among the chromosomes than I previously thought. It seems like everything except a few exceptions (Y, 8, 14, 16, 22) are pretty diverse. Every chromosome (besides the previously mentioned exceptions) has some oncogenes and some tumor suppressors, and they all have some genes for cell survival and some for cell fate. Only 6 chromosomes have genome maintenance but it is still spread out. Something our group noticed was that all genome maintenance genes are also tumor suppressor which makes sense because they have to work together to fix the DNA and suppress the cell divination. This is probably the biggest thing I learned from this activity, I had previously thought that the genes where much more specialized.
Paragraph 2: Another thing I learned is that the different types of cancer (at least Melanoma) aren’t very restricted on the genes they attack or what chromosomes or functions or anything. In my Melanoma group we didn’t notice many patterns, the genes where not located on the same chromosomes, most of the genes where not the same except 2 of us had BRAF and two had MLL3, the same number of genes where not affected (two of us had 5 affected and 1 of us had 4 affected), and last but not least there was no pattern in the functions, we had mixed of cell fate and cell survival and two had maintenance but it didn’t seem to matter. We came to the conclusion that this means it is way easier to get cancer than we thought. This makes it seem very wide spread and not very specific to a certain gene or chromosome. Yes some came up more but it was all so randomly distributed that it seems widespread and harder to prevent because you don’t know where its attacking you cell.
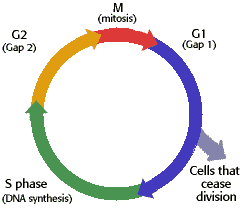
Paragraph 3: One thing I learned not from this activity but from the week was about the cycle a cell goes through to divide and all the checkpoints and things. There are 4 stages: G1, S, G2, and M. G1 is the period of growth before the DNA is replicated, S is the period where the DNA is replicated, then G2 where the cell grows more now that there is new DNA and it prepares for division, and then there is M which is Mitosis and the cell divides.
Paragraph 4: The two main things that surprised me was how diverse the genes and chromosomes where and how few genes there where. I expected the chromosomes to be much more specialized, it is weird to me that all the different types of genes are seemingly spread of evenly. I also expected there to be so many more and it seems like there just aren’t that many different genes.
Paragraph 5 (question): How easy is it to get cancer and are some genes / chromosomes better at fighting it than others?

